Effect of protease (trypsin) on the flippase activity of spinach ER... | Download Scientific Diagram

Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. | Semantic Scholar

Progress in Our Understanding of Wzx Flippase for Translocation of Bacterial Membrane Lipid-Linked Oligosaccharide. - Abstract - Europe PMC

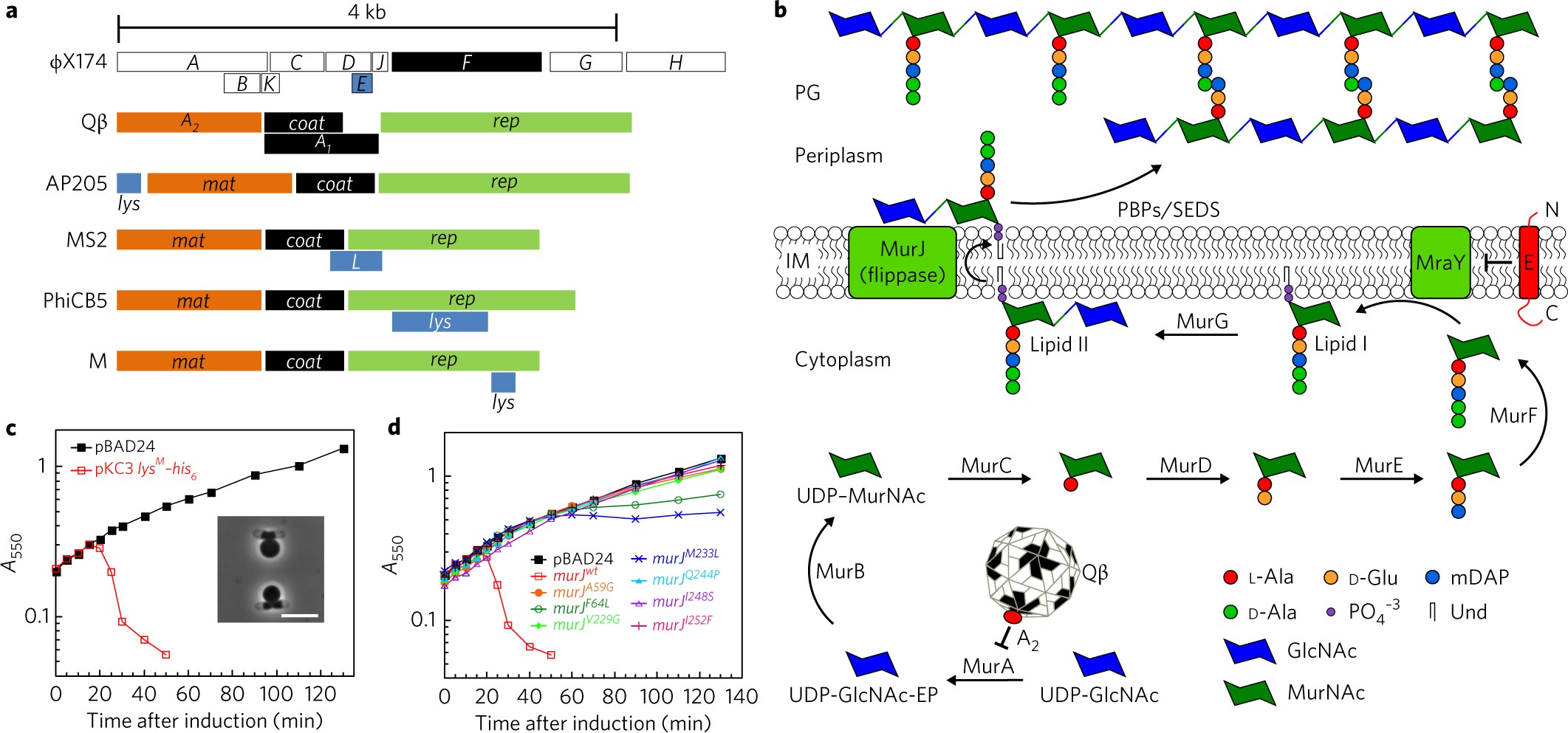
Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling | Journal of the American Chemical Society

Crystal structure of the MOP flippase MurJ in an inward-facing conformation. - Abstract - Europe PMC

Loss of specificity variants of WzxC suggest that substrate recognition is coupled with transporter opening in MOP‐family flippases - Sham - 2018 - Molecular Microbiology - Wiley Online Library

Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling | Journal of the American Chemical Society

Progress in Our Understanding of Wzx Flippase for Translocation of Bacterial Membrane Lipid-Linked Oligosaccharide | Journal of Bacteriology

Crystal structure of the lipid flippase MurJ in a “squeezed” form distinct from its inward- and outward-facing forms - ScienceDirect

Crystal structure of the MOP flippase MurJ in an inward-facing conformation | Nature Structural & Molecular Biology

Detection of Transport Intermediates in the Peptidoglycan Flippase MurJ Identifies Residues Essential for Conformational Cycling | Journal of the American Chemical Society
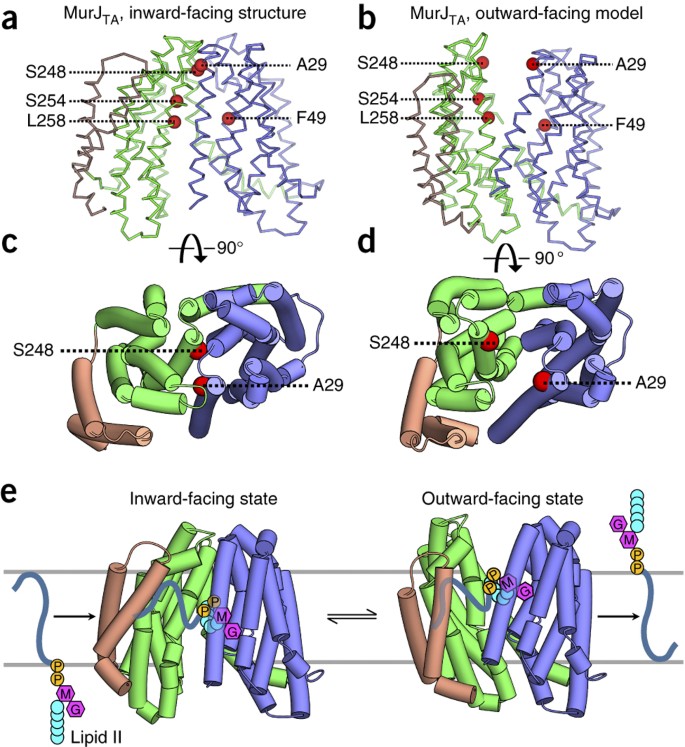
Crystal structure of the MOP flippase MurJ in an inward-facing conformation | Nature Structural & Molecular Biology